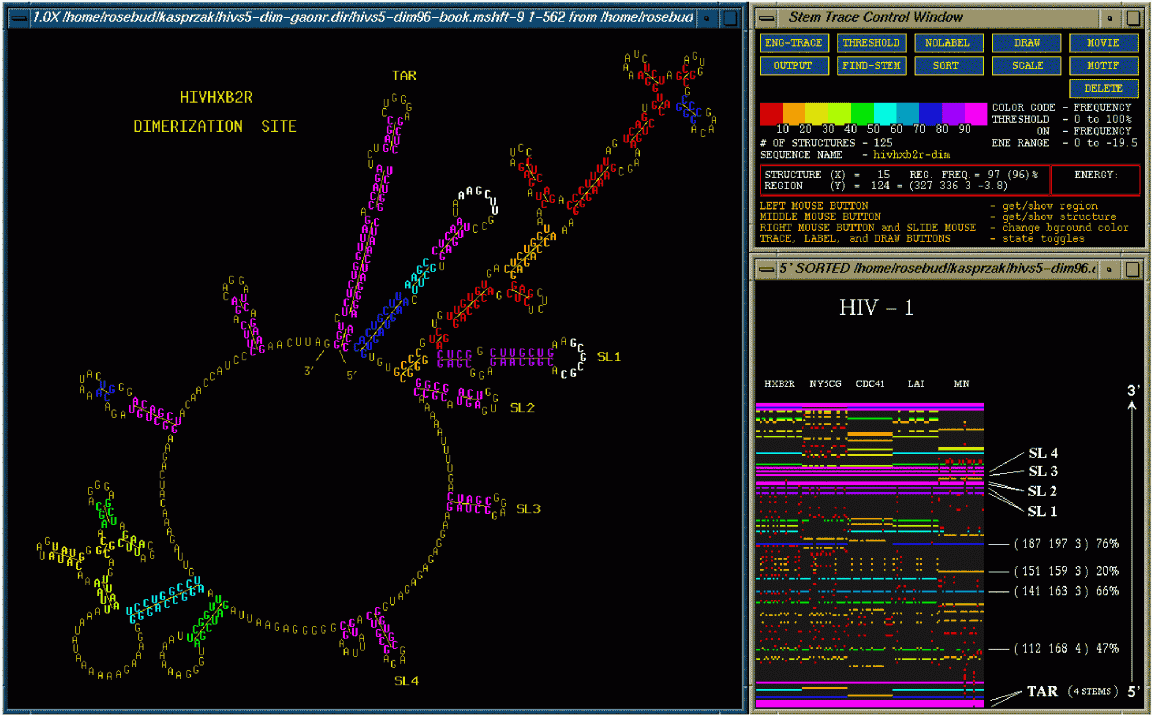
The above figure shows the Stem Trace Control Window (upper right), the trace window (lower right), and an associated 2D structure drawing, showing one of the folded structures with the stems color-coded to reflect their frequency in the stem trace shown. In this case the above figure shows the folds of 5 different strains of HIV-1. Each strain was folded 25 times by the GA (as it is a non-deterministic algorithm) to obtain a consensus. This consensus is depicted in the stem trace. Sequence alignment is performed to make sure that the stems are properly matched up. The stems are color-coded depending on the frequency of their appearance in different foldings of the different HIV-1 strains. Magenta means that the stem appeared in over 90% of the structures while red means that the stem appeared in less then 10% of the structures. The control window (upper right) contains function buttons dedicated to displaying and manipulating the stem data currently pointed to in the trace window. The user may interact with the elements in the stem trace using these function buttons to retrieve various types of information concerning the individual structures presented. These buttons allow further information extraction and visualization. The trace window (lower right) shows all the predicted, color coded stems, position-sorted from 5' to 3'. The different strains of HIV-1 are labeled at the top and the 25 color coded foldings are lined up beneath. This allows one to visualize where the stems are identical in the solution spaces of a family of sequences.
For more information see (Kasprzak and Shapiro 1999).