Our research deals with the development and use of computational methodologies for the prediction and analysis of RNA structure. Our objective is to understand the structural characteristics of the control mechanisms that are inherent in many RNAs. Moreover, we are beginning to apply our knowledge to the development of methodologies for RNA nanobiology. Our research may be broken down into the following major areas:
Development of RNA Structure Prediction and Analysis Software
We have developed a massively parallel genetic algorithm, MPGAfold, for RNA structure prediction. This algorithm is capable of predicting H-type pseudoknots and to determine potentially significant intermediate structures along a folding pathway. In addition, we can visualize and interact with the structure maturation process in real-time by the MPGAfold Visualizer. This is a Java based application which extends the functionality of MPGAfold. When MPGAfold is used in conjunction with the MPGAfold Visualizer, all operating and running conditions of MPGAfold are delegated to the MPGAfold Visualizer.
In addition to and in conjunction with the visualizer, RNA structure analysis is performed with help of a comprehensive computer workbench, StructureLab, which permits the examination of very large solution spaces of structures that may be generated by various folding programs. It offers algorithms for structural comparisons, motif searching and structural visualization. STRUCTURELAB is a distributed computing environment. Remote RNA structure analysis algorithms can be run on several different computer architectures that run the Unix or Linux operating systems. The results from these computations can then be further analyzed by the workbench.
Another aspect of our research involves determining RNA structure from sequence alignment. Two projects (KNetFold and CorreLogo) analyze sequence alignments with respect to potential conserved RNA secondary structure elements. One key concept is to use compensatory base changes in an alignment. KNetFold also uses thermodynamic data and conservation of Watson-Crick base pairs in a machine learning algorithm to make base pair predictions.
Application of the Computational Methodologies for Analysis of RNA Structure Folding Dynamics
We have applied the above metioned methodologies to understand the RNA secondary structure folding dynamics of several biological systems. These include for example HIV, rotavirus, pstv, hdv and hok/sok. Of particular interest in most of these cases is the characterization of multiple functional conformational states.
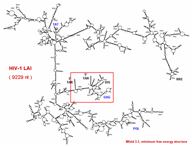
Modeling and Characterizing the Three-Dimensional Atomic Level Structure of RNA
We apply modeling methodologies and molecular dynamics to understand the atomic level functional details of RNA molecules. Particular attention has been paid to a portion of telomerase, the core central domain of the 16S rRNA and the HIV-1 kissing loop complex. Examples are illustrated here.
RNA Nanobiology
We are developing algorithms for the design of RNA based nanodevices. RNA nanoconstructs can serve as building blocks and scaffolds to assemble more complex functional objects. These objects can potentially deliver functional groups (RNA, peptides or otherwise) to cells controlling cell genetics, cell death, or the visualization of delivery pathways. Towards this endeavor we have been characterizing RNA motifs and developing RNA nano design tools.